
BendixLab - Biophotonics & Mechanobiology
Dive into the intersection of biology and physics. Our mission is to decode how cells convert force, shape, and motion into biological function. To achieve this, we work at the crossroads of theoretical physics, molecular and cell biology, medicine, nanoscience, and plant science, creating a shared quantitative framework that enables truly interdisciplinary collaboration.
We investigate how cells use physics to organize their cell surface proteins through phase separation and geometry and how cells sense mechanical signal from their environment. Explore our findings in GPCR mechanosensing, cell surface dynamics, repair and organization using interesting new experimental methods.
We are a group of enthusiastic and creative scientists interested in understanding physical properties of biological systems at the single molecule to whole cell level.
The group has a strong interdisciplinary profile with a number of close collaborators in biology, nanoscience, medicine, chemistry and theory (see staff section for more details).
This is a short film about our research. Watch the full film here.
We have dual optical trapping platform combined with a confocal microscope which allows cutting edge experiments to be performed. Also, we have super resolution microscopy based on the STORM method. For imaging larger specimens, like embryos, we have light sheet microscopy which facilitates fast and low photo-toxicity imaging. Microscopic physical properties of cells are quantified using an optical trap as a force sensor/actuator whereas whole cell properties can be explored using the cell deformation cytometry.
Additionally, we have an extensive expertise in working with model membrane systems and also isolated plasma membrane systems containing the membrane proteins from the cell.
We are located at the Niels Bohr Institute in the Niels Bohr Building at the University of Copenhagen.
Funding our research

Research at the interface between biophotonics and mechanobiology
We explore how cells respond to physical stimuli such as membrane curvature and mechanical forces. We investigate how optical tweezers can be used to study mechanobiology, including mechanosensing and interactions at the single-molecule level. Read on to learn more about our projects and results.
Mechanosensing by GPCR
In collaboration with the Biomedical Institute at UCPH we investigate how G-protein-coupled receptors (GPCRs) (a major family of cell-surface receptors) respond to mechanical forces. This includes advanced techniques to manipulate and measure forces at the single-molecule level, providing insight into signaling mechanisms.
Our research contributes to a deeper understanding of how cells sense and respond to their mechanical environment.
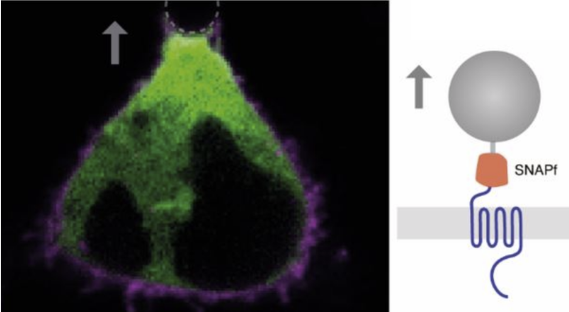
By using optical tweezers or other mechanical tools on cells we apply local or global mechanical stresses to the cells. The biochemical response is monitored using miniGs which bind to GPCRs upon activation.
This work is part of an interdisciplinary consortium called GPCRmec, funded by the Novo Nordisk Foundation, with multiple academic partners.
Membrane curvatures
Membrane curvature plays a crucial role in cellular processes.
We investigate how proteins generate and sense or respond to membrane curvature, which is essential for processes such as vesicle formation, cell migration, and filopodia formation. This involves the use of advanced microscopy techniques in parallel with biophysical methods such as optical tweezers to deform membranes into high membrane curvatures.
Review: Emerging Topics in Life sci. (2023) Ruhoff et al.
Review: BioChemical Society Trans. (2023), Ruhoff et al.
Soft Matter (2021) Florentsen et al.
ACS Central Science (2020), Larsen et al.
ACS Nano (2019) Moreno-Pescador
Nature Chemical Biology (2017), Rosholm et al.
Soft Matter (2015), Barooji et al.
Sci. Rep. (2013), Ramesh et al.
Filopodia pulling and twisting dynamics
We discovered a new way that filopodia work. Filopodia are thin, finger-like protrusions that cells use as “feelers” to sense and grab their surroundings. They are found on many cell types and are especially common on moving immune cells and cancer cells where they facilitate invasion.
We found that filopodia can rotate because their inner support structure—made of actin fibers—can twist like a rope. This was the first clear evidence that a bundled set of actin fibers can twist in this way. We also suggested a simple physical explanation for how the twisting happens.
This twisting matters because it can help the cell pull on things it touches—similar to how a twisted rubber band tends to tighten and pull as it relaxes.
- Nature Communications, 2022, Leijnse et al.
- PNAS 2015, Leijnse et al.
- Review in TCB : Ruhoff et al. 2025
- Review in Cytoskeleton 2015 Leijnse et al.
Thermoplasmonics for biology
We have been pioneers in the field of plasmonics in biology. We have quantified and mapped out the optical heating from various nanostructures both theoretically and experimentally. Subsequently, we have used thermoplasmonics—tiny metal structures that can be heated with light—to do very precise manipulation or “microsurgery” on cells or soft materials.

With this method, we can make controlled, local damage to the cell surface or even the membrane around the nucleus (in collaboration with the Cancer Institute). This lets us study how cells sense and repair injuries, which happen often when cells squeeze and move through tissue—especially immune cells and cancer cells.
We also use the same light-based heating tool for bioengineering: for example, to merge (fuse) cell membranes with synthetic membranes, or to locally “switch” membranes into a different physical state in a specific spot (a bit like changing a small patch of butter from solid to soft without affecting the rest).
- Nano LettersReview (2024), Ruhoff et al.
- JOVE (2024), Danielsen et al.
- Nano Letters (2023) Ruhoff et al.
- Nanoscale (2022), Moreno-Pescador et al.
- Chemical Reviews(2019), Jauffred et al.
- ROPP (2017), Bahadori et al.
- Sci. Rep. (2016), Norregaard et al.
- Nano Letters (2015), Rørvig-Lund et al.
- ASC Nano (2010), Bendix et al.
- Nano Letters (2005), Hansen et al.
Single Molecule Biophysics
Using advanced optical trapping (4 trap system from Lumicks) together with microfluidics and confocal imaging, we explore the DNA-protein interactions taking place during replication.
In collaboration with researchers from molecular biology at UCPH we investigate an array of transcription factors and proteins which help organize the genome.
Plant Biophysics
In collaboration with the Staffan Persson Lab (Plant Science, UCPH), Guillermo Pescador (UCPH) and Alexander Rohrbach (Freiburg), we have developed a new label-free microscopy approach that detects intracellular dynamics in plant cells through light scattering. Using this method, we can reveal and quantify previously hidden diffusion dynamics of intracellular vesicles in plant roots.
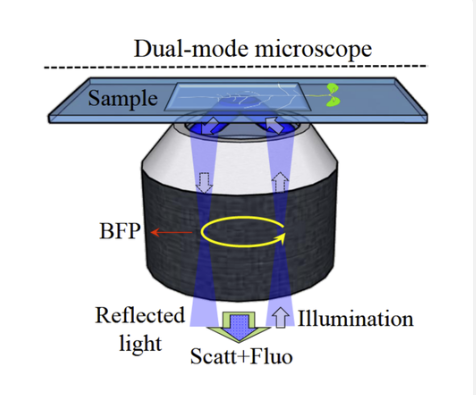
We also study the biophysics of the plant cell surface using optical trapping and thermoplasmonics as manipulation tools. In particular, we investigate how membrane phase behavior, tension and membrane curvature shape the lateral distribution of proteins.
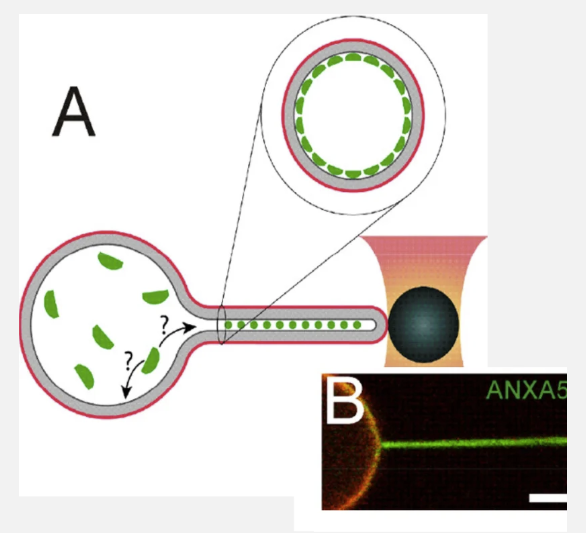
Cell membrane repair and biophysics of annexins
In collaboration with Jesper Nylandsted at the Danish Research Center we have investigated how cells repair surface lesions by recruiting different types of annexins. We have locally punctures living cells while monitoring recruitment of fluorescent annexins to the site of damage.
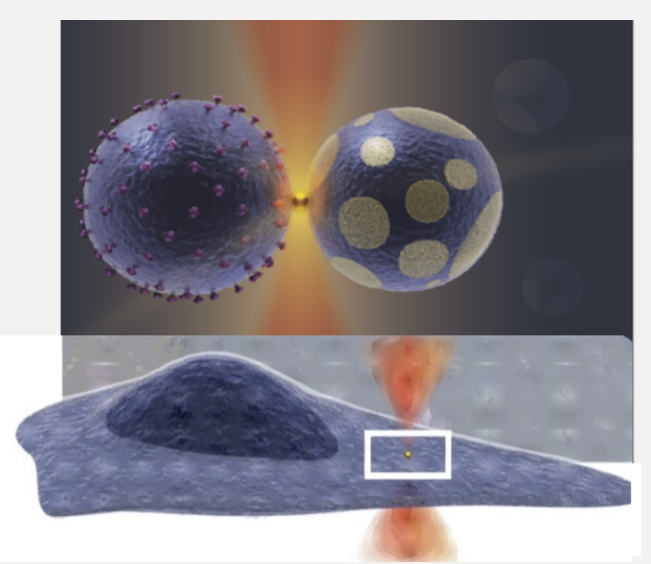
To gain biophysical insight into the mechanism of annexins we have made in vitro studies using membrane vesicles with encapsulated annexins. These vesicles were optically shape modulated or punctures to understand the reaction of annexins to shape and to formation membrane holes.
Thermoplasmonics is currently being used to study the repair mechanisms of nuclear membranes as well.
Her is an overview of our main experimental facilities which our lab members and collaborators have free access to.
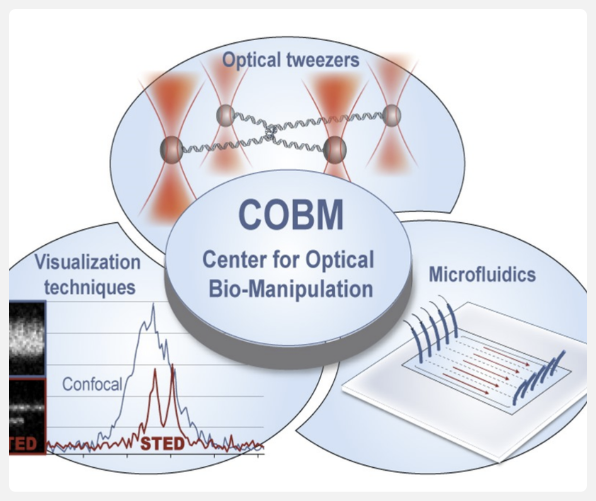
Optical trapping infrastructure
We offer experimental services and collaborative opportunities for researchers who want to explore optical trapping—from single-molecule biophysics to experiments with cells.
Our platform is the newest and most advanced optical trapping system currently available. It combines four independently controlled optical traps with confocal and super-resolution microscopy, integrated microfluidics, and a highly user-friendly interface—enabling a broad range of high-impact experiments with precise control and flexibility.

Fluorescent Microscopy
Our lab has three confocal microscopes and STORM superresolution microscopy for imaging everything from cells to single molecules. These systems are coupled to other systems such as optical tweezers or mechanical imaging (Brillouin Microscopy).

We have all typical laser wavelengths for excitation and photomultiplier tubes for collection of signals.
We offer committed collaborations of alternatively a pay-per-use basis.
Please contact us for more information.
Link to infrastructure: Center for Optical Bio-Manipulation (COBM)
Brillouin Imaging
Want to image mechanical properties of matter? Then try Brillouin Microscopy which is a microscopy combining acoustics with laser scattering to achieve images showing the mechanical stiffness of your material.
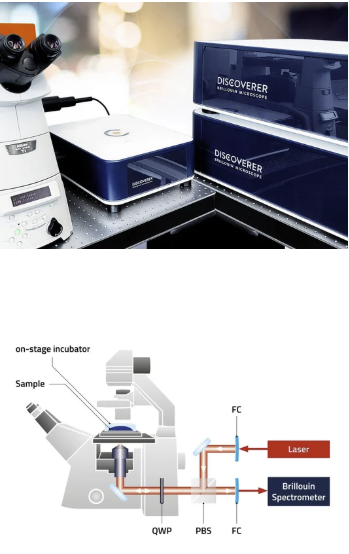
We use this for imaging living cells. This equipment is shared with Doostmohammadi Lab at the Niels Bohr Academy, UCPH
Scattering/interferometric microscopy
Scattering and interferometric microscopy allows us to detect nanoscopic objects with no fluorescence. This label free technology permits fast imaging at millisecond timescale or can be used for long term imaging of plant cells. Plant cells are particularly interesting for interferometric imaging due to the plant wall barrier which prevents use of intracellular markers.
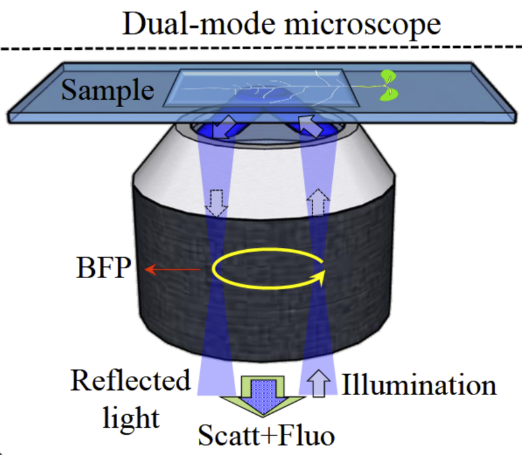
Our microscope is a Rotating Optical Coherence Scattering (ROCS) microscope which has a lateral resolution of down to 150nm and axial resolution down to 10 nm.
Mechanical perturbation
We also modulate cell shapes by mechanical action. Squeezing cells between a glass coverslip and an adjustable piston allows accurate compression of cells with 1 micrometer axial resolution. This way we can test cell response to confined conditions which are often experienced by living cells in vivo.

Periodic stretching of a cell substrate is an alternative mechanical method for deformation of cells. Cells can be fixed just after being stretched to detection any molecular or structural responses due to the stretching.
The cell squeezer can operate while imaging cells using confocal microscopy
We bridge other scientific fields with experimental biophysics. Our collaborators work in diverse fields such as molecular, theoretical, medical and plant science.
GPCRmec
This consortium includes collaborators from the medical institute and Chemistry. The goal is to understand how mechanics influences g-protein coupled receptor signaling. Financed by a Novo interdisciplinary synergy grant.
SEECLEAR
An unconventional collaboration between theoretical and experimental physics and plant science. Our goal is to understand formatiomn of the secondary cell wall. Financed by a Novo exploratory synergy grant.
Active collaborations
Signe Mathiasen
Biomedical Institute, UCPH
Staffan Persson
Plant Science, UCPH
Weria Pezheskian
Theoretical physics, Niels Bohr Institute, UCPH
Julien Duxin
Biotech Research & Innovation Centre, UCPH
Pétur Heiðarsson
Department of Biology, UCPH
Jesper Nylandsted
Danish Research Center, UCPH
Karen Martinez
Chemistry, UCPH
Mette Rosenkilde
Biomedical Institute, UCPH
Kalina Haas
INRAE, Paris, France
Jakub Sedzinski
Novo Nordisk Foundation
Center for Stem Cell Medicine, reNEW, UCPH
We have many different projects for Master's students and also occasional projects for undergraduate students.
This is a video of Victoria Ruhoff explaining her project about biophysics of virus proteins (Danish).
Cellular repair system investigated by laser based nano-surgery
When cells migrate in the body they often experience membrane ruptures which results in excessive calcium flowing into the cell. This can be lethal unless the membrane is sealed within milliseconds. Cells have a built-in surface repair kit which is activated by calcium influx and hence can allow the cell to self-heal within seconds after injury. Cancer cells are extremely efficient in repairing their surface since they have an increased expression of various annexin proteins which are thought to be the major proteins involved in the membrane repair.

The project will involve testing the membrane repair system in cells by using thermoplasmonics to inflict a nanoscopic hole in the membrane. This will be done by irradiating a plasmonic gold nanostructure placed on the surface of the cell. Confocal microscopy and super-resolution microscopy will be used to monitor the recruitment of various annexins which are labeled with fluorescent proteins. The overall aims are to i) investigate whether invasive cancer cells are more efficient in dealing with thermoplasmonic ruptures than non-invasive cells ii) investigate the role of several different annexins in the repair process including other proteins like ESCRT and actin.
The project is a collaboration with Kræftens Bekæmpelse samt Syddansk Universitet.
Contact: Poul Martin Bendix, bendix@nbi.dk
Patterning in large bacterial communities
 In nature, bacteria actively search for a surface to form larger communities, i.e., biofilm, with extended cooperativity and defense. We know that sectors with low genetic diversity form within the colony, even among cells of similar fitness. This self-organization of microbial cell communities is the result of genetic drift in complex interplay with evolution, competition, and cooperation.
In nature, bacteria actively search for a surface to form larger communities, i.e., biofilm, with extended cooperativity and defense. We know that sectors with low genetic diversity form within the colony, even among cells of similar fitness. This self-organization of microbial cell communities is the result of genetic drift in complex interplay with evolution, competition, and cooperation.
We offer various projects to explore pattern formation by growing bacteria both in vivo and in silico. We believe the close interplay between theory and experiments will provide a more complete understanding of cooperation and competition among cells in larger communities. We aim to point out general features of growth pattern, which can be generalized in wider class of systems. In the long term, we may draw parallels to mammalian cell systems, where patterning is crucial for example in embryonic development.
Possible subprojects include:
Colony shape
The relation between the individual cell shape and the colony shape
This project combines theory and experiments depending on your interests. Experimentally, the project can include bacterial cell culture, colony growth, and advanced fluorescence microscopy. Theoretically, we plan to first simulate an individual cell-based model where the particles grow, divide, and interact through mechanical force. Depending on the development of the project, simplified lattice models or partial differential equation-based models can also be used.
Supervisors: Liselotte Jauffred & Namiko Mitarai
Signe Mathiasen
Biomedical Institute, UCPH
Staffan Persson
Plant Science, UCPH
Weria Pezheskian
Theoretical physics, Niels Bohr Institute, UCPH
Julien Duxin
Biotech Research & Innovation Centre, UCPH
Pétur Heiðarsson
Department of Biology, UCPH
Jesper Nylandsted
Danish Research Center, UCPH
Karen Martinez
Chemistry, UCPH
Mette Rosenkilde
Biomedical Institute, UCPH
Kalina Haas
INRAE, Paris, France
Jakub Sedzinski
Novo Nordisk Foundation
Center for Stem Cell Medicine, reNEW, UCPH
Coming to the Experimental Biophysics with an external grant
The Experimental Biophysics supports researchers' applications to international and Danish public and private funding agencies.
If you come to the group with your own funding, you will become a full member of the team with office space, access to IT and admin support, laboratories, and you are expected to contribute to the scientific and social life in the group.
Examples of agencies funding research in biophysics in Denmark:
-
Independent Research Fund Denmark | Natural Sciences (FNU) and Medical Sciences (FSS) - public; postdocs, phd's, research groups, mobility
-
European Research Council (ERC) - international; research groups
-
European Commission Horizon 2020 - international; postdocs, mobility
-
Novo Nordisk Foundation - private; postdocs, research groups, mobility
- Lundbeck Foundation - private; postdocs, phd's, research groups, mobility
.. and several others. You may also consider applying for funding from private or national agencies in your home country, and name the Optical Tweezers Group at the Niels Bohr Institute as your host institution.
Procedure
Contact Group Leader, Assoc. Prof. Poul Martin Bendix, if you are considering applying for external funding.
Please contact us:
- at least 8 weeks in advance of the application deadline in the case of individual post doc stipends and smaller projects (i.e., 1-3 years, under DKK 3M);
- at least 3 months in advance of the application deadline in the case of larger projects.
The group's faculty will review your request to apply. The Niels Bohr Institute may be able to offer some co-funding in the case of larger projects/grants. A needs assessment for equipment may also be conducted, to ensure that the Institute can adequately house your project and the expected personnel associated with it.
If approved, we can provide considerable support for the application process, for example including budget development, text about the host institution, review of application's science case, and contact with the funding agency if needed. The Faculty of Science Research Funding Office at the University of Copenhagen also provides support for applications.
An application budget is developed online using the University's application tool and must be approved by the group leader and the Niels Bohr Institute at least one month before the application deadline. The budget process also facilitates the conversation about possible co-financing from the Institute or Center, laboratory set-up if needed, and any HR issues that need to be addressed.
 Poul Martin Bendix, Group leader
Poul Martin Bendix, Group leader
E-mail: bendix@nbi.ku.dk
Tel: +45 35325251 or +45 61602454
Niels Bohr Building, Jagtvej 132
2200 Copenhagen N.
Staff
| Name | Title | Job responsibilities | |
|---|---|---|---|
| Search in Name | Search in Title | Search in Job responsibilities | |
| Bendix, Pól Martin | Associate Professor | Associate Professor, Group Leader |
|
| Farhangi Barooji, Younes | Academic Staff |
|
|
| Hamel Ascanio, Luis Eduardo | PhD Student |
|
|
| Mesi, Elsa | PhD Fellow |
|
|
| Moreno Pescador, Guillermo Sergio | Guest Researcher |
|
Master/undergraduate students
| Navn | Titel | Foto | |
|---|---|---|---|
| Natálie Palková | INTERACT PhD student from Duxin Lab | natalie.palkova @bric.ku.dk |
 |
| Ciara Dooley | Master Student | rhp186@alumni.ku.dk | |
| Noëlle Klasner | Master Student | noelle.klasner@gmail.com | |
| Laura Isabella Pultz Henriksen | Bachelor Student | laura.henriksen@nbi.ku.dk | |
| Konrad Skovmand Olsen | Voluntary student | kol@adm.ku.dk |
News on Biocomplexity and Biophysics


Physicists develop modeling software to diagnose serious diseases







