Biocomplexity and Biophysics
Biocomplexity is a cutting-edge area of research between physics and biology. By using the principles and methods of physics one can explore the living nature and biological phenomena.
"More is different" by P. W. Anderson emphasizes emergent phenomena on a wide range of scales in nature.
In BioComplexity, we continuously explore the diversity of complex phenomena in biological, physical and social systems, including pattern formation, complex and chaotic dynamics, fluid dynamics, game theory, networks and econophysics.
We build on our strength of using physics approaches to suggest and perform experiments and models of living systems. The systems range from proteins and gene regulation to larger-scale collective spatiotemporal structure formation. The research is often a collaboration between physicists, biologists, medical doctors, and nanoscientists.
Biocomplexity section consists of researchers with a variety of interests. Fascinated by diverse phenomena in biological and complex systems, we are tackling the research topics theoretically, computationally, and experimentally.
We have a biocomplexity seminar series, with open seminars every 1-2 weeks, normally on Wednesdays. In addition, researchers flexibly organizes seminars and discussions.
We welcome students interested in working in this fascinating, cutting edge research area. We attract a large number of master students to write a thesis in bio- and/or complex system research projects.
The Niels Bohr institute has many M.Sc. and PhD students. The students are closely attached to the research groups and supervisor, and have many social activities for International and Danish students.
If you are interested in studying Biophysics or Biocomplexity, consider looking at these pages:
General information about the education, application, living in Denmark etc.
Biocomplexity:

Biological Network and Complex systems (Center for Models of Life (CMOL)): We use methods from physics to develop understanding of living systems. We model regulation of living systems aiming to understand the strategies of gene regulation and dynamics of self organization, with a focus on simple conceptual and quantitative models. Our research tackles diversity of biological systems from epigenetics to emergence of complex communities.
Universal Biology group: The Universal biology group at the Niels Bohr Institute aims to unveil universal characteristics in life systems and universal laws in adaptation, development, and evolution, with the spirit of physics (in particular, statistical physics and dynamical systems), by noting consistency between hierarchical levels – molecules, cells, organisms, and ecosystem
Read more about Universal Biology Group >>
Biophotonics & Mechanobiology:

BendixLab: We investigate the physical properties of living cells by a number of advanced optical techniques and methods from eg. nanoscience. Living cells are extremely dynamic, and for a physicist, the complex cell can be characterized as a living material. We examine the cells by combining optical microscopy with optical manipulation.
Read more about Experimental Biophysics >>

Membranes Lab: Using physical principles and methods, the group 'Biophysics - Membranes' explores biological phenomena and works to clarify how proteins and chemical substances pass through both biological and artificial membranes. The physical theory of an electrical circuit can, for example, describe how signal transports around the brain.

Uni-Bio lab: from models to unifying concepts in Biology:
We use mathematical models and quantitative experiments to investigate how individual cells and cellular populations rapidly adapt to changes in the environment. A few examples include Asymmetric Damage Segregation (bacteria), Early differentiation of the embryo, receptor adaptation; excitable dynamics in inflammation (islets).
Center for Models of Life >> |
|
NanoHeal (JM/CMOL)The application of stress to multiphase solid-liquid systems often results in morphological instability. For example, a surface of a stressed solid in contact with its solution corrugates and develops parallel grooves. The instability is triggered by deformation mechanisms operating across multiple scales and it is thermodynamically controlled by spatial gradients in the chemical potential. |
|
FlowTrans (JM/CMOL)The research themes of FLOWTRANS relate to the characterization and the understanding of fluid flow and chemical reaction within rocks and granular media. This has become an ever-increasing problem in Earth Sciences, Physics, and in many industrial applications, including CO2 sequestration, hydrocarbon migration, ore deposit development, and radioactive waste disposal. |
|
Epigenetics (CMOL)Cells carry information handed down from their ancestors and are able to pass on information to their descendants. In many cases this memory" is epigenetic - not stored in the DNA sequence - allowing cells with identical DNA to maintain distinct properties. |
|
Collective behavior in cell communities (CMOL)Biological signaling and regulation of cell growth and state may extend across space, combining local action of some molecules with the spatially extended action of molecules that are designed to diffuse more freely. |
|
Self organization of Diversity: Evolution, species, diseases, development (CMOL)At all scales, biology presents an astounding diversity of discrete states or species, which coexist over time-scales much longer than the characteristic times of the underlying degrees of freedom. These degrees of freedom ranges from protein to population numbers. |
|
Regulatory dynamics and feedbacks (CMOL)Protein production in living cells is a huge process, which depends on external conditions, stress, and also coupled to the ability of genetically identical cells to behave differently. The transcription and translation machinery of cells are regulated on both large and small scale, invoking physiological stress signals, as well as specifically acting transcription and translation regulators. |
|
Experimental Biophysics >> |
|
BIOPHYSICS OF THE CELL SURFACEIn this group of projects our aim is to understand how cells dynamically organize their cell surface and perform all the functions associated with the surface of the cell. Examples include remodeling of the plasma membrane, cell - cell interactions, lateral redistribution of proteins in the membrane and membrane repair. We study these physical phenomena like filopodia dynamics, cell surface repair by annexins, membrane curvature sensing by proteins, lipid order in plasma membranes and resulting protein segregation into distinct lipid phases. To explore these phenomena we use confocal imaging, super resolution (STORM), optical trapping, plasmonic heating and membrane manipulation tools. Specific questions include: What kind of dynamics do filopodia exhibit?The cell surface contains mechanically active antennae-like structures called filopodia. Our lab has contributed significantly to the understanding of the biophysical properties of these structures. We have found that they have the ability to rotate, pull and form helical buckles – all at the same time. These properties have been measured quantitatively using optical trapping in conjunction with 3D confocal imaging (Lejinse et al. PNAS, 2015). Funding: Villum Foundation (2012-2015) How do cells cope with plasma membrane ruptures?Currently, we are exploring the mechanism of plasma membrane repair in living cells. Cells are punctured using thermoplasmonics which allows nanoscopic holes to be formed in the cell membrane. Several annexins are involved in the cell repair and we are doing parallel imaging and puncturing to reveal the mechanism of the cell surface repair machinery. In parallel we study the biophysical properties (curvature affinity and mechanical effects of protein binding) of the same proteins in models systems like GUVs and plasma membrane vesicles. Collaborators: Robert Daniels (FDA), Jesper Nylandsted (Kræftens Bekæmpelse), Adam Cohen Simonsen (SDU), Himanshu Khandelia (SDU) Funding: Novonordisk synergy grant (2019-2022). How do proteins affect membrane mechanics?Proteins which sense and generate membrane curvature are often found to mechanically stiffen membranes. We have studied the mechanical effects of proteins bound within nanotubes (Ramesh et al. Sci. Rep., 2013 & Barooji et al. Sci. Rep., 2016). The mechanical effect is typically correlated with the ability of the protein to oligomerize. Do proteins sense lipid order and membrane curvature?Both peripheral and integral proteins are found to sense the lipid ordering in plasma membranes. Influenza virus protein Neuraminidase was found to sort exclusively into lipid disordered domains in isolated plasma membranes (Moreno et al. ACS Nano, 2019). Membrane curvature also constitutes another mechanism of lateral organization of proteins. We have shown that F-BAR (Ramesh et al. Sci. Rep., 2013), I-BAR (Barooji et al. Sci. Rep., 2016), Annexin V (Moreno et al. ACS Nano, 2019) and GPCR (Rosholm et al. Nat. Chem. Biol., 2017) effectively sense membrane curvatures and the sensing correlates with the molecular shape of the protein. Collaborators: Robert Daniels (FDA), Jesper Nylandsted (Kræftens Bekæmpelse), Dimitrios Stamou (Nanoscience Center, KU), Szabolcs Semsey (SNIRP BIOME) Funding: Villum Foundation (2012-2015) and Sapere Aude DFF (2015-2019). Can the plasma membrane of living cell be isolated with all its content intact?During recent years we have focused on development of a method for isolation of the plasma membrane from living cells. This method is highly attractive since it allows us to investigate the plasma membrane proteins in their natural membrane environment, and in absence of the cellular skeleton and other organelles. The isolated membrane forms a spherical vesicle containing the proteins from the mother cell and also internal cytosolic proteins. Importantly, these proteins preserve their correct orientation in the membrane which is in striking contrast to reconstitution of proteins in membranes. This work was published in ACS Nano in 2019 (Moreno et al.) where we showed the feasibility of using these vesicles for optical manipulations and for studying lipid phase behaviors. Collaborators: Robert Daniels (Food and Drug Administration, USA), Jesper Nylandsted (Kræftens Bekæmpelse), Szabolcs Semsey (SNIRP BIOME). Funding: DFF Sapere Aude (2015-2019) and Novonordisk synergy grant (2019-2022). |
|
|
Funding: Villum Foundation (2012-2015) and Sapere Aude DFF (2015-2019) and Novonordisk synergy grant (2019-2022). |
|
|
More about the research in the Experimental Biophysics group >> |
|
Uni-Bio-Lab >>
|
|
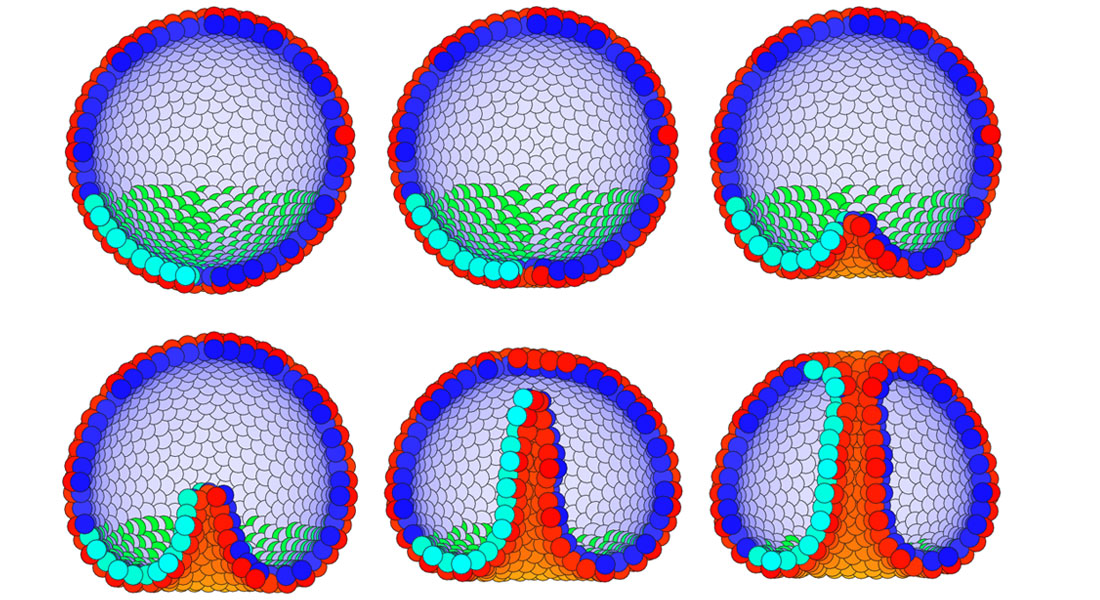
Models of embryos and organs (Uni-Bio-Lab)Finances: Stemphys, DNRF, Period: 2015-2020 Staff and Students: Silas Boye Nissen, Alexander Nielsen, Julius Bier Kirkegaard. Contact Person: Ala Trusina Papers:1) Theoretical tool bridging cell polarities with development of robust morphologies, SB Nissen, S Rønhild, A Trusina, K Sneppen, Elife 7, e38407, 2017 2) Stochastic priming and spatial cues orchestrate heterogeneous clonal contribution to mouse pancreas organogenesis HL Larsen, L Martín-Coll, AV Nielsen, CVE Wright, A Trusina, YH Kim, Anne Grapin-Botton, Nature communications 8 (1), 605 3) Four simple rules that are sufficient to generate the mammalian blastocyst. / Nissen, Silas Boye; Perera Pérez, Marta; Martin Gonzalez, Javier; Morgani, Sophie Maria Christina; Jensen, Mogens Høgh; Sneppen, Kim; Brickman, Joshua Mark; Trusina, Ala. I: PLOS Biology, Vol. 15, Nr. 7, e2000737, 12.07.2017, s. 1-30 |
Bente Markussen, Secretary
Niels Bohr Building, Biokomplexity and Biophysics
Jagtvej 132, 2200 København N.
Phone: +45 353-35845
Mobil: +45 23839875
E-mail: markussen@nbi.ku.dk
Namiko Mitarai, Head of section
Jagtvej 132, 2200 København N.
Phone: +45 35 32 54 02
Email: mitarai@nbi.ku.dk
Staff
| Name | Title | Phone | |
|---|---|---|---|
| Search in Name | Search in Title | Search in Phone | |
| Abbasova, Nigar | PhD Fellow | +4535323591 | |
| Agafonova, Alisa | PhD Student | ||
| Alonso, Albert | Guest Researcher | +4535325426 | |
| Andersen, Simon Guldager | PhD Fellow | ||
| Bendix, Pól Martin | Associate Professor | +4535325251 | |
| Berard, Adrien Raymond | PhD Fellow | +4535335191 | |
| Bergamaschi, Laura | PhD Fellow | +4535322371 | |
| Bonn, Lasse Frederik | PhD Fellow | +4535329685 | |
| Bravo Vidal, Adrià | PhD Fellow | +4535332071 | |
| Brown, Stanley | Associate Professor Emeritus | ||
| Bölsterli, Robin Vincent | PhD Fellow | +4535336732 | |
| Cherian, Philip | Postdoc | +4535327068 | |
| Doostmohammadi, Amin | Associate Professor - Promotion Programme | +4535331427 | |
| Doshi, Harsh | PhD Fellow | +4535332381 | |
| Ehrmann, Anja | Postdoc | ||
| Eilersen, Andreas Thomas | Guest Researcher | ||
| Ellegaard, Clive Sigurd | Associate Professor Emeritus | +4520576305 | |
| Farhangi Barooji, Younes | Academic Staff | +4535330067 | |
| Geiger, Beatrice Jacqueline | PhD Fellow | +4535332757 | |
| Giorgi, Alessandro | PhD Fellow | +4535322433 | |
| Hamel Ascanio, Luis Eduardo | PhD Student | ||
| Heimburg, Thomas Rainer | Professor | +4535325389 | |
| Heltberg, Mathias Spliid | Assistant Professor | +4535327500 | |
| Hertz, John | Emeritus | +4535325235 | |
| Hvid, Ulrik | PhD Fellow | +4535329088 | |
| Ichii, Shunsuke | Guest Researcher | ||
| Ikeda, Yutaro | Guest Researcher | ||
| Jauffred, Liselotte | Guest Researcher | +4535324857 | |
| Jensen, Mogens Høgh | Professor | +4535325371 | |
| Jessen, Emilie | PhD Fellow | +4535334408 | |
| Kaneko, Kunihiko | Professor | +4535326343 | |
| Kirkegaard, Julius Bier | Assistant Professor | +4540938516 | |
| Klahn, Mathias | Postdoc | +4535332982 | |
| Knudsen, Teresa Emmilie Toudal | Postdoc | +4535331216 | |
| Kristensen, Lukas Hvemon | PhD Fellow | +4535326649 | |
| Levinsen, Mogens | Associate Professor Emeritus | +4535325295 | |
| Lindahl, Fanny Isabell | PhD Fellow | +4535337861 | |
| Lucchetti, Alessandra | Postdoc | +4535337508 | |
| Lund, Oskar Struer | PhD Student | +4535330269 | |
| Ma, Tianxiang | PhD Fellow | +4535328035 | |
| Marantos, Anastasios | Guest Researcher | +4535330159 | |
| Mesi, Elsa | PhD Fellow | +4535321768 | |
| Mitarai, Namiko | Associate Professor | +4535325402 | |
| Moreno Pescador, Guillermo Sergio | Guest Researcher | +4535332635 | |
| Nakamura, Yoshiyuki | Visiting PhD Student | ||
| Nguyen, Thu Trang | Visiting Student | ||
| Nielsen, Bjarke Frost | Postdoc | ||
| Nielsen, Malthe Skytte Nordentoft | PhD Student | ||
| Nishide, Ryosuke | Postdoc | ||
| Noronha, Riz Fernando | PhD Fellow | +4535328970 | |
| Novev, Yavor Kirilov | Guest Researcher | +4535334916 | |
| Pedersen, Martin Cramer | Associate Professor | +4535334852 | |
| Pezeshkian, Weria | Assistant Professor | +4535324714 | |
| Plugers, Davey Antonino | PhD Fellow | +4535331604 | |
| Ratnasinghe, Brian Damith | PhD Fellow | +4535337618 | |
| Rocchetti, Michela | PhD Fellow | +4535325120 | |
| Sargado, Juan Michael Uy Villanueva | Postdoc | +4535334017 | |
| Schauser, Jakob Hallundbæk | PhD Fellow | +4535329993 | |
| Skjegstad, Lars Erik Johnsen | PhD Fellow | +4535329126 | |
| Sneppen, Kim | Professor | +4535325352 | |
| Stillits, Andreas | PhD Student | +4535329562 | |
| Thomsen, Brage Haldor | Research Assistant | +4535335042 | |
| Trusina, Ala | Associate Professor | +4521288647 | |
| Trzaska, Jakub | PhD Fellow | +4535328187 | |
| Utecht, Sebastian Øllegaard | Research Assistant | ||
| Venkatesh, Varun | PhD Fellow | +4535329217 | |
| Vieira Flauzino, João Vitor | PhD Fellow | +4535328869 | |
| Xu, Xiaochan | Guest Researcher | +4535326306 | |
| Yadav, Anjali | PhD Fellow | +4535329456 | |
| Yu, Chengyou | PhD Marie Curie | +4535327972 | |
| de Graaf Sousa, Niels | PhD Fellow | ||
| van den Berg, Nathánaël | Research Assistant | +4535331929 |
News on Biocomplexity and Biophysics


Physicists develop modeling software to diagnose serious diseases