 Center for Models of Life
Center for Models of Life
In the Center for Models of Life, CMOL, we use methods from physics to develop understanding of living systems. We model regulation of living systems aiming to understand the strategies of gene regulation and dynamics of self organization, with a focus on simple conceptual and quantitative models.
Our research tackles diversity of biological systems from biological development to emergence of complex communities.
The Center for Models of Life started as a center of excellence initiative under the Danish National Research Foundation in 2005.
The idea of the center was spawned at the interface between physics and biology, with inspiration from the many new quantitative tools that was developed in the biological sciences. This opened for a new and more number based approach to understand living systems, in particular on the genetic and epigenetic level.
As the center matured, we have expanded the research directions, and members of the center are now engaged in a number of projects that extend from epigenetics to stem cells and from virus to microbiomes. Diversity in itself has become a major inspiration for our activities, as we repeatedly see that "more is different".
In the Center for Models of Life, we develop physics models that allow us to build a quantitative understanding of computation and communication in biological systems.
Networks are common themes in complex systems research, ranging from the regulation of actions in the simplest viruses to ecological and social systems. Our approach is to model regulative networks of living systems, including analysis of the physical mechanism of interactions, the dynamics of information transfer along signaling pathways, as well as modeling slower evolutionary changes in the networks. Our ambition is to quantify life as a dynamic and evolving system composed of networks of interacting parts with a history.
Biological Development
Biological Development is the dynamical process of making an embryo and forming the vast diversity of cells and shapes of organs within an organism. This involves cellular decisions and self-assembly of many cells into organs.

Despite continual renewal and damages, a multicellular organism is able to assemble and maintain its complex morphology. How is this robustness compatible with the complexity and diversity of living forms? Inspired by the progressive role of apical-basal and planar cell polarity in development, we propose that stability, complexity, and diversity are emergent properties in populations of proliferating polarized cells.
Collective behavior in cell communities
Biological signaling and regulation of cell growth and state may extend across space, combining local action of some molecules with the spatially extended action of molecules that are designed to diffuse more freely.
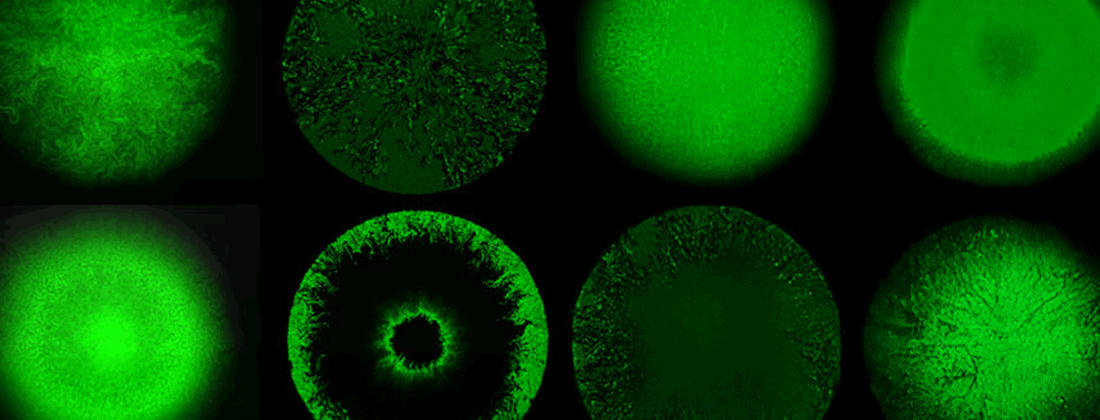
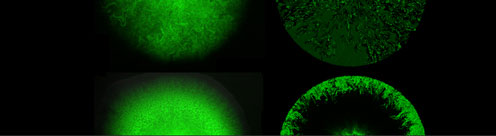
Combining production with secretion, or cellular growth with mutual inhibition of growth, one may obtain a large variety of both frozen and dynamic patterns including pattern as advanced as embryo formation and somite formation in vertebrates. On the population level, spatio-temporal patterns can also be seen in growing bacterial, respectively yeast colonies. In our center, we work with a variety of these spatio-temporal model systems, spanning from plaque formation of phages as they spread among growing bacterial cultures.
Self organization of Diversity: Evolution, species, diseases, development
At all scales, biology presents an astounding diversity of discrete states or species, which coexist over time-scales much longer than the characteristic times of the underlying degrees of freedom. These degrees of freedom ranges from protein to population numbers.

On the sub-cellular scale, molecular competition and positive feedback acting on short time scales maintain cells in a variety of specialized states, setting the foundation for complexity of multicellular organisms. On larger scales, competition in turn selects for ecosystems where different species coexist over long intervals of time.
This general pattern calls for research with a focus on diversity, and how competition can act as its ``engine". Statistical mechanics of complex systems provides a framework for studying universality of collective and cooperative phenomena, usually assuming the simplicity of identical agents.
Regulatory dynamics and feedbacks
Protein production in living cells is a huge process, which depends on external conditions, stress, and also coupled to the ability of genetically identical cells to behave differently.
The transcription and translation machinery of cells are regulated on both large and small scale, invoking physiological stress signals, as well as specifically acting transcription and translation regulators.
The networks of such regulations defines a feedback, which in turn regulates decisions in living cells.
In some cases, e.g. in development of an animal or in the lysis-lysogeny decisions of temperate phages, a gene regulatory system decides which of a few possible states to take. Such decisions involves positive feedbacks and will be exposed to stochastic fluctuations at molecular level.
When cell need to maintain life functions and homeostasis, then negative feedback is invoked. Combinations of positive and negative feedback are opens for spatial patterns and transmission of information across space.
In collaboration with biological laboratories we consider a number of such gene regulatory systems and how they guide cells and tissues to work. Current examples include (p)ppGpp and ribosome traffic in E.coli, iron and sugar metabolism networks, phage decision networks, toxin-antitoxin systems, as well as the mammalian systems of inflammation response (NFkappaB).
Epigenetics
Cells carry information handed down from their ancestors and are able to pass on information to their descendants. In many cases this ``memory" is epigenetic -- not stored in the DNA sequence -- allowing cells with identical DNA to maintain distinct properties.
Epigenetic cell memory implies alternative states that are stable over time and are inherited through cell division. Mechanism for epigenetic cell memory invokes positive feedback loops, either on genes with mutual repression or, for eukaryotes, through recruited modifications of DNA bound nucleosomes.
We explore the rules for this feedback, through a combined experimental (With Genevive Thon) and theoretical modeling, with aim to determine how stable bistability can be combined with locality for the spreading of the epigenetic states.
Another epigenetic mark is methylation respectively demethylation of CpG islands in mammals. When relatively short segment of DNA with high density of CpG sites are positioned close to a promoter, they regulate the activity of this promoter. We explore how such systems can be actively maintained, assuming that they themselves contribute to the feedback that maintain them persistently in a either methylated or a unmethylated state.
You can either click on a person at the bottom of the page and see their publications, or you can see a complete list of publications for the entire section for Biocomplexity.
- Mogens H. Jensen, Professor
- Kim Sneppen, Professor
- Kunihiko Kaneko, Professor
- Joachim Mathiesen, Professor
- Namiko Mitarai, Associate Professor
- Ala Trusina, Associate Professor
- Mathias Spliid Heltberg, Assistant Professor
Each PI in our group works independently on distinct research topics. If you have inquiries—such as those regarding postdoc or PhD positions, internships, or research collaborations—please review the individual research profiles and reach out directly to the PI whose work aligns with your interests.
Staff
| Name | Title | Phone | |
|---|---|---|---|
| Search in Name | Search in Title | Search in Phone | |
| Agafonova, Alisa | PhD Student | ||
| Bergamaschi, Laura | PhD Fellow | +4535322371 | |
| Brown, Stanley | Associate Professor Emeritus | ||
| Ehrmann, Anja | Postdoc | ||
| Heltberg, Mathias Spliid | Assistant Professor | +4535327500 | |
| Hvid, Ulrik | PhD Fellow | +4535329088 | |
| Jensen, Mogens Høgh | Professor | +4535325371 | |
| Kaneko, Kunihiko | Professor | +4535326343 | |
| Kirkegaard, Julius Bier | Assistant Professor | +4540938516 | |
| Klahn, Mathias | Postdoc | +4535332982 | |
| Knudsen, Teresa Emmilie Toudal | Postdoc | +4535331216 | |
| Lund, Oskar Struer | PhD Student | +4535330269 | |
| Marantos, Anastasios | Guest Researcher | +4535330159 | |
| Mitarai, Namiko | Associate Professor | +4535325402 | |
| Nakamura, Yoshiyuki | Visiting PhD Student | ||
| Nishide, Ryosuke | Postdoc | ||
| Noronha, Riz Fernando | PhD Fellow | +4535328970 | |
| Plugers, Davey Antonino | PhD Fellow | +4535331604 | |
| Sargado, Juan Michael Uy Villanueva | Postdoc | +4535334017 | |
| Schauser, Jakob Hallundbæk | PhD Fellow | +4535329993 | |
| Sneppen, Kim | Professor | +4535325352 | |
| Stillits, Andreas | PhD Student | +4535329562 | |
| Trusina, Ala | Associate Professor | +4521288647 | |
| Yadav, Anjali | PhD Fellow | +4535329456 |
